You'll receive the latest updates on new standards, guidelines, and educational resources, as well as expert insights to help enhance your laboratory's performance and compliance.
Applying Susceptibility Interpretations to the Candida parapsilosis Species Complex

Shawn R. Lockhart, Centers for Disease Control and Prevention, Atlanta, GA
As DNA sequence- and MALDI-TOF-based identification of yeasts becomes widespread, we have learned much about the prevalence of cryptic species within species complexes. One of the more common Candida species, Candida parapsilosis, is part of a three species complex including C. orthopsilosis and C. metapsilosis. Both C. orthopsilosis and C. metapsilosis are cryptic species. From a worldwide surveillance program collected between 2006 and 2016, the C. parapsilosis species complex isolates were comprised of 95.5% C. parapsilosis, 3.2% C. orthopsilosis, and 1.3% C. metapsilosis. 1 A conundrum that arises in the clinical laboratory is that there are separate epidemiological cutoff values (ECVs) for each of the three species,2 but there are only breakpoints for C. parapsilosis. 3 How should the laboratory report results of antifungal susceptibility tests of C. parapsilosis complex isolates?
If a laboratory is able to perform species identification within the species complex:
- C. parapsilosis breakpoints should only be used for C. parapsilosis sensu stricto isolates.
- For isolates identified as C. metapsilosis or C. orthopsilosis, the species-specific ECVs should be used with a comment that ECVs are not a predictor of clinical outcome but may help identify non-wild type isolates. Other suggestions of interpretive comments regarding ECVs are available within the CLSI M59 document.2
If a laboratory is able to identify C. parapsilosis but cannot distinguish the other species within the complex, the C. parapsilosis breakpoints can be used. When the original C. parapsilosis breakpoints were set, it is likely that a low number of cryptic species within this complex were included. If laboratories cannot distinguish between the complex and are applying the C. parapsilosis breakpoints, it may be prudent to include a comment such as the following: “Candida parapsilosis is part of a species complex but the other species cannot be distinguished by the procedures used in this laboratory. While the prevalence of these other species is low, the predictive clinical response when applying the C. parapsilosis breakpoints used here to them is unknown.”
For antifungals with no breakpoints or ECVs of C. parapsilosis or other members of the species complex, such as flucytosine, no interpretation should be reported. It is not acceptable to use an ECV from a closely related species for interpretation.
Table 1. Candida parapsilosis breakpoints and species specific epidemiological cutoff values for the C. parapsilosis species complex2,3
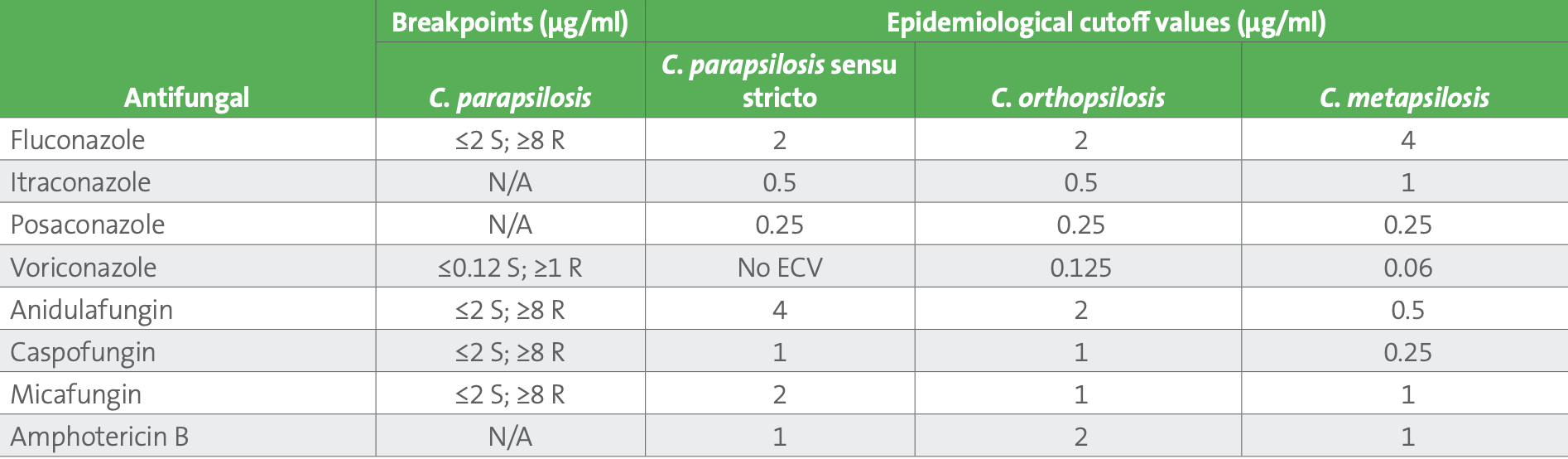
Abbreviations: N/A, not available; R, resistant; S, susceptible.
References:
1 Pfaller MA, DJ Diekema, JD Turnidge, M Castanheira, and RN Jones (2019) Twenty years of the SENTRY antifungal surveillance program: results for Candida species from 1997-2016. Open Forum Infect Dis. 6(Suppl 1):S79-S94.
2 CLSI. Performance Standards for Antifungal Susceptibility Testing of Yeasts, 2nd Ed. CLSI supplement M60. Wayne, PA: Clinical and Laboratory Standards Institute; 2020.
3 CLSI. Epidemiological Cutoff Values for Antifungal Susceptibility Testing, 3rd Ed. CLSI supplement M59. Wayne, PA: Clinical and Laboratory Standards Institute; 2020.
Full AST News Update July 2020