You'll receive the latest updates on new standards, guidelines, and educational resources, as well as expert insights to help enhance your laboratory's performance and compliance.
AST News Update June 2022: Case Studies

What’s Wrong with This Picture? Case 1
Stella Antonara, Ohio Health, Columbus, OH
Lars F. Westblade, Weill Cornell Medicine, New York, NY
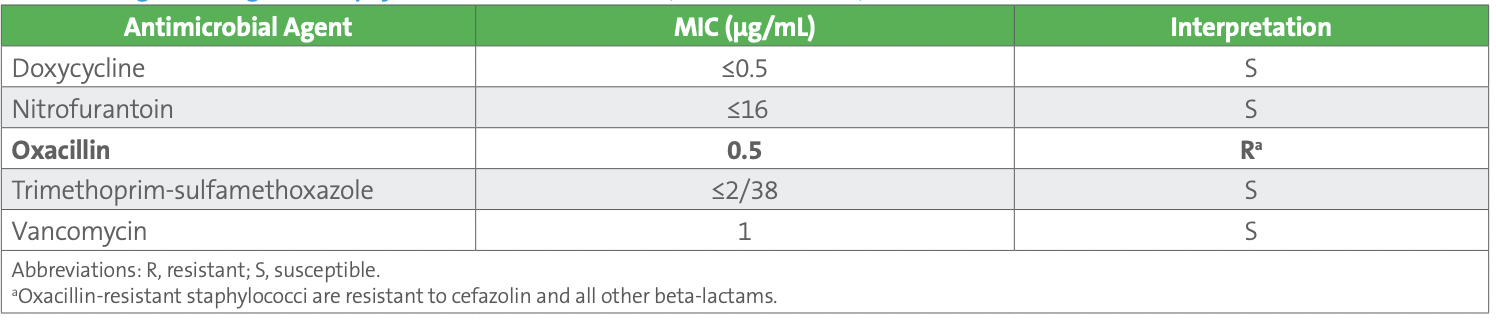
Case 1: Coagulase-negative Staphylococcus spp. An 86-year-old man with low blood pressure and elevated heart and respiratory rates presented to the emergency department with complaints of painful urination, fever, and chills. His white blood cell count was 1,000/mm3 and he had elevated lactic acid levels. The patient was diagnosed with urosepsis, urine and blood cultures were ordered, and the patient was admitted. The urine culture was positive with >100,000 CFU/mL coagulase-negative staphylococci (CoNS) (pure culture) with antimicrobial susceptibility test (AST) results obtained from a commercial AST device (see Table 1).
Table 1. Coagulase-negative Staphylococcus - Urine Culture (Released Results)
Both blood culture sets were positive with gram-positive cocci in clusters within 24 hours of collection. A molecular test was performed on the positive blood culture broth from one of the aerobic bottles and was positive for Staphylococcus species and negative for the mecA gene. Subcultures from both sets of blood cultures revealed CoNS, based upon spot tests, and identified as Staphylococcus hominis. The blood isolate was preliminarily reported as “methicillin (oxacillin)-susceptible S. hominis, further susceptibilities to follow.” AST was performed on the isolate recovered from the blood culture bottle used for molecular testing, again using the commercial AST device. Results are shown in Table 2.
Table 2. Staphylococcus hominis - Blood Culture (Unconfirmed Results)
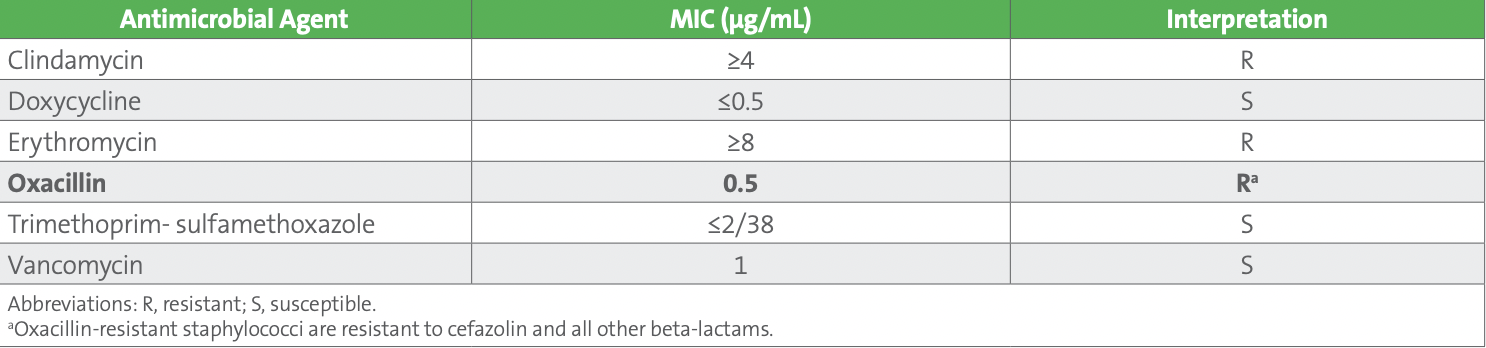
The technologist reviewing the results noticed that the oxacillin MIC interpretation of “Resistant” from the blood isolate did not agree with the negative result for mecA on the molecular panel performed on the positive blood culture broth. What‘s wrong with this picture?
Solution to Case 1: The urine culture isolate was identified as S. hominis using matrix-assisted laser desorption/ionization timeof-flight mass spectrometry (MALDI-TOF MS), but, according to current laboratory practice, CoNS other than Staphylococcus saprophyticus are not reported to the species level. Instead, the isolate was initially reported as “coagulase-negative Staphylococcus spp.” but was subsequently reported to the species level, given the clinical picture of urosepsis and the presence of the isolate in pure culture. In contrast, CoNS isolated from blood cultures are identified and reported to the species level. The blood culture isolate was identified as S. hominis.
The AST results for the blood culture and urine culture isolates were identical. These included clindamycin and erythromycin susceptibility results that were suppressed on the urine AST report. These agents should not be routinely reported on urine isolates as they are not effective in treating urinary tract infections. However, the oxacillin MIC interpretation for the blood isolate was inconsistent with the molecular panel results for which the mecA gene was not detected.
The following steps were taken to troubleshoot the discrepancy between the oxacillin AST and mecA results:
1. All subculture plates (including the AST purity plate) from the positive blood culture were reviewed for the presence of additional colony morphologies.
– Subcultures were pure and only one colony type was noted, which was confirmed by MALDI-TOF MS to be S. hominis.
2. The molecular panel was run again on the positive blood culture broth (within the manufacturer-approved time after positive signal).
– Identical results were obtained: Staphylococcus species detected, mecA not detected.
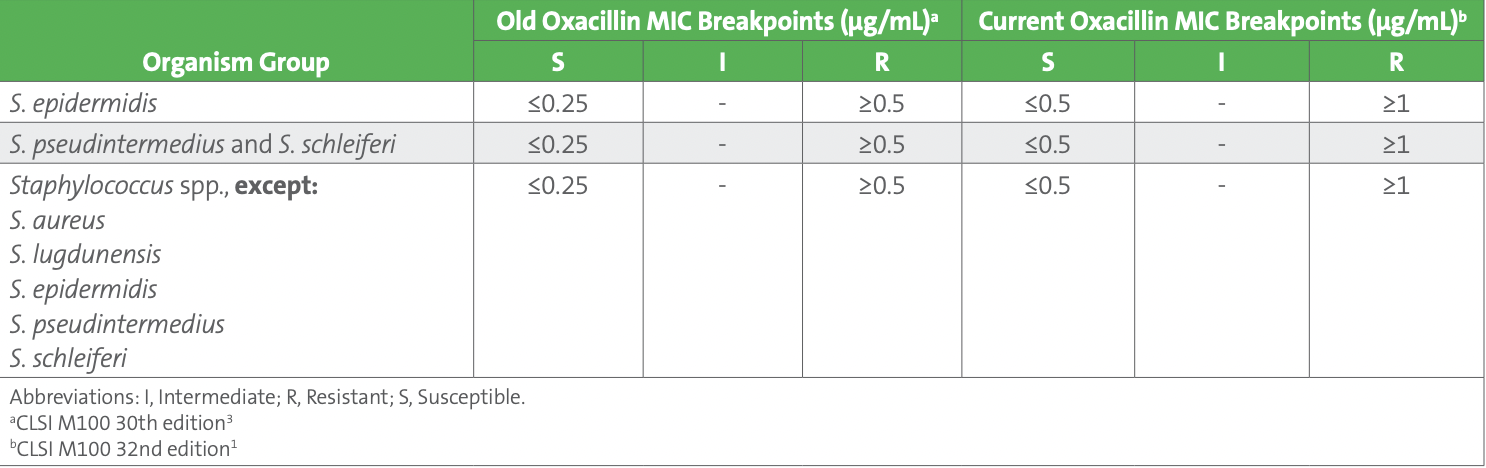
Subsequently, one of the technologists recalled that oxacillin MIC breakpoints were recently updated by CLSI in the M100 31st edition published in 2021.1 The oxacillin susceptible breakpoint for all Staphylococcus species, except Staphylococcus aureus and Staphylococcus lugdunensis, was changed from ≤0.25 µg/mL to ≤0.5 µg/mL (see Table 3). This update was based on recent studies that demonstrated an oxacillin breakpoint of ≤0.5 µg/mL for susceptible correlated better with the absence of the mecA gene, especially for Staphylococcus capitis, Staphylococcus haemolyticus, S. hominis, and Staphylococcus warneri isolates. Consequently, there are fewer false resistant results (major errors) with these updated breakpoints.2
Table 3. Oxacillin Breakpoints for Staphylococcus Species Other Than Staphylococcus aureus and Staphylococcus lugdunensis
The updated oxacillin breakpoints improve performance of oxacillin MIC tests with Staphylococcus spp. other than S. aureus and S. lugdunensis, however they are not perfect. This is particularly true for those species for which species-specific oxacillin breakpoints have not been set (ie, Staphylococcus spp. excluding the following: S. aureus, S. lugdunensis, Staphylococcus epidermidis, Staphylococcus pseudintermedius, and Staphylococcus schleiferi; OR, coagulase-negative Staphylococcus spp., which have not been identified to the species level). In fact, no phenotypic test is highly reliable for these species when oxacillin MICs are 1-2 µg/mL. Therefore, CLSI suggests testing for mecA or PBP2a for isolates with oxacillin MICs of 1-2 µg/mL from serious infections included in the category Staphylococcus spp., except: S. aureus, S. lugdunensis, S. epidermidis, S. pseudintermedius, and S. schleiferi. “Isolates that test mecA or PBP2a negative should be reported as methicillin (oxacillin) susceptible.” 1
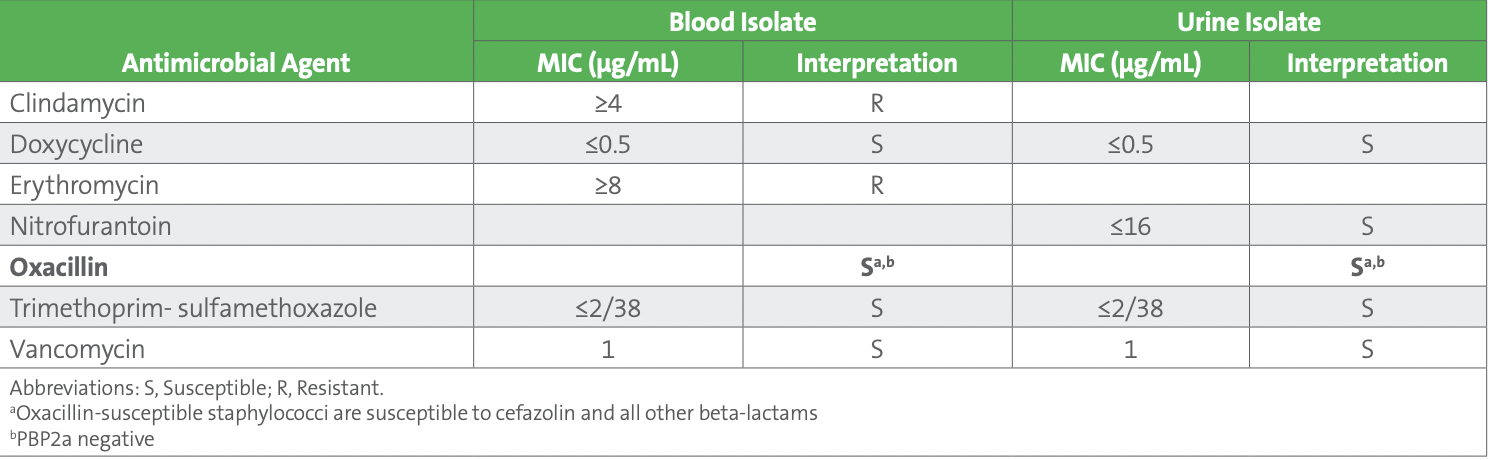
As part of the troubleshooting process, the urine and blood isolates were tested with PBP2a and results were negative, confirming the lack of mecA previously obtained by molecular testing of blood. Considering the updated breakpoints, and the negative mecA and PBP2a results, both the blood and urine isolates were reported as oxacillin susceptible (see Table 4).
Table 4. Staphylococcus hominis - Blood Culture (Released Results) and Urine Culture (Corrected Results)
Upon review of the breakpoints applied by the commercial AST device, the laboratory discovered that the system followed the prior CLSI oxacillin MIC breakpoints (ie, M100 30th Ed). Therefore, oxacillin MIC results were not released, as the laboratory had not yet officially validated the updated breakpoints. At this point, FDA has not approved the updated oxacillin breakpoints recommended by CLSI (FDA oxacillin breakpoints can be found on the FDA website4 ). It is important to remember that commercial manufacturers must use FDA breakpoints in their AST systems. However, laboratories can implement updated CLSI breakpoints, including the new oxacillin breakpoints, on their commercial AST system following performance of a validation.
This article describes a case wherein the laboratory applied the term “coagulase-negative Staphylococcus spp.,” but determination of oxacillin resistance relied on species level identification of the organism. If phenotypic susceptibility testing and reporting are undertaken for oxacillin and/or cefoxitin, species level identification of CoNS must be performed. The decision to implement the updated staphylococcal oxacillin breakpoints should be made with the antimicrobial stewardship team. Laboratories should consider including mecA or PBP2a testing on all isolates of coagulase-negative staphylococci from serious infections (eg, prosthetic joints, blood cultures) for those isolates with oxacillin MICs of 1-2 µg/mL.
References
1. CLSI. Performance Standards for Antimicrobial Susceptibility Testing. 32nd ed. CLSI supplement M100. Malvern, PA: Clinical and Laboratory Standards Institute; 2022.
2. Humphries RM, Magnano P, Burnham CA, et al. Evaluation of surrogate tests for the presence of mecA-mediated methicillin resistance in Staphylococcus capitis, Staphylococcus haemolyticus, Staphylococcus hominis, and Staphylococcus warneri. J Clin Microbiol. 2020;59(1):e02290-20. doi:10.1128/JCM.02290-20.
3. CLSI. Performance Standards for Antimicrobial Susceptibility Testing. 30th ed. CLSI supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute; 2020.
4. https://www.fda.gov/drugs/development-resources/antibacterial-susceptibility-test-interpretive-criteria. Accessed May 4, 2022.
What’s Wrong with This Picture? Case 2
Lars F. Westblade, Weill Cornell Medicine, New York, NY
Daniel D. Rhoads, Cleveland Clinic, Cleveland, OH
Stella Antonara, OhioHealth, Columbus, OH
Case 2: Serratia marcescens. Direct specimen Gram stain of a tracheal aspirate revealed a moderate quantity of gram-negative bacilli and rare polymorphonuclear cells. In culture, many Stenotrophomonas maltophilia, Pseudomonas aeruginosa, and Serratia marcescens were recovered and antimicrobial susceptibility testing (AST) was performed using an automated system. The AST results for the S. maltophilia and P. aeruginosa isolates were unremarkable. The S. marcescens AST profile is tabulated below (see Table 1). What’s wrong with this picture?
Table 1. Serratia marcescens
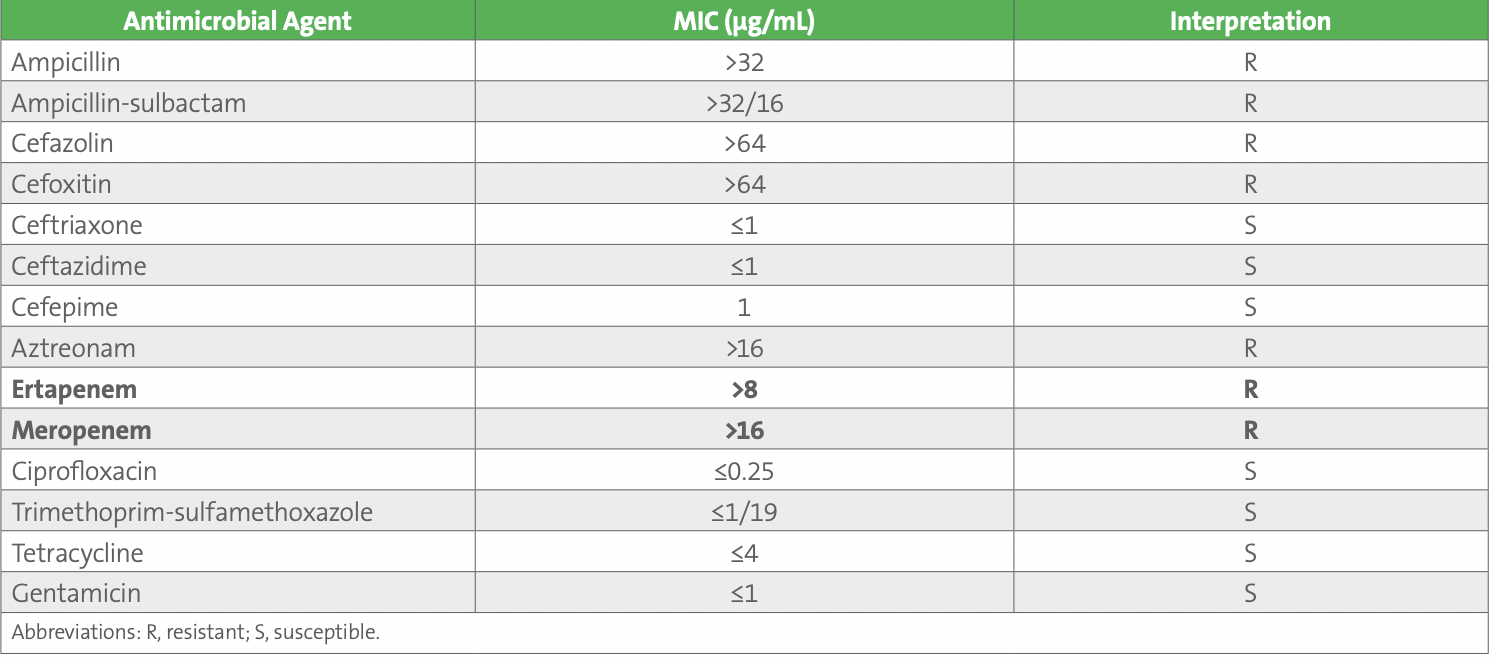
Solution to Case 2: The S. marcescens isolate’s observed susceptibility to extended-spectrum cephalosporins (ceftriaxone, ceftazidime, and cefepime) in the setting of carbapenem (ertapenem and meropenem) resistance is unusual. Possible explanations for this observation are listed below:
1. Contamination of the AST panel. In this scenario, contamination of the AST panel with a meropenem-resistant isolate.
2. Technical error during AST panel set up.
3. Unusual or rare resistance mechanism. In this case, an unusual or rare β-lactam resistance mechanism.
The points listed above serve as a good starting framework to explain possible AST problems. Due to the unusual AST results, they were not released, and the case was raised to the AST technical specialist for further counsel. They suggested the following steps to investigate the root cause of the unusual AST profile. First, the purity plate was closely examined. It was pure for a single organism and the identity of the isolate was confirmed to be S. marcescens by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS), demonstrating the AST panel was unlikely contaminated with a carbapenemresistant species. Second, the results were discussed with the technologist who performed the testing. The technologist did not recall any issues with the automated instrument or the panels the previous day and had many years of experience with the automated AST system, suggesting technical error was unlikely on the part of the technologist. Furthermore, no issues with QC or the platform itself had been recorded for the past month implying instrument and/or panel error was improbable. Finally, given observed resistance to meropenem, the isolate was assayed using a commercial immunoassay for IMP, KPC, NDM, OXA-48-type, and VIM enzymes. It was negative for all five targets. Based on the performing laboratory’s testing algorithm, all carbapenemresistant isolates that test negative using the carbapenemase detection immunoassay are tested using a phenotypic (enzymatic) carbapenemase detection test (CDT) (Carba NP), which was positive.
It was determined the S. marcescens isolate was likely producing an S. marcescens enzyme (SME). The AST was repeated using the same method and the initial results were confirmed. The AST results were released as they tested, as is recommended by the Clinical and Laboratory Standards Institute (CLSI).1 SMEs are chromosomally-encoded serine carbapenemases that have only been observed in S. marcescens. 2,3 Four SME variants (SME-1 to SME-4) have been described, and they are resistant to carbapenems but susceptible to extended-spectrum cephalosporins,2,3 as observed in our case. Of note, a similar AST profile (susceptible to extended-spectrum cephalosporins and resistant to carbapenems) can be observed for OXA-48-type-producing Enterobacterales.3 The first description of SME-producing S. marcescens was from England in 1982 and they have remained relatively uncommon since that initial report, with most cases observed in the United States.3,4 Due to their location on the chromosome, transmission of blaSME (the gene encoding SME carbapenemases) is considered to be less of an infection control risk compared to other carbapenemases (eg, IMP, KPC, NDM, OXA-48-type, and VIM) where mobile genetic element (eg, plasmid)-mediated transmission is possible. Nevertheless, measures must be taken to contain SME-producing S. marcescens to prevent outbreaks resulting from clonal dissemination.2 Interestingly, mobilization of blaSME between S. marcescens isolates has been proposed via an excised circular intermediate of a cryptic prophage genomic island: S. marcescens genomic isolate 1-1, suggesting horizontal transfer between S. marcescens isolates is possible and that isolates harboring blaSME could be more of an infection control risk than previously appreciated.4
The detection of SME-producing S. marcescens isolates can be difficult because of their susceptibility to extended-spectrum cephalosporins and the omission of blaSME from CDTs designed to detect carbapenemase genes.2,5 With the exception of the modified Hodge test, which is no longer endorsed by CLSI, most phenotypic CDTs readily detect SME-mediated carbapenemase activity.1,4,5,6 It has been proposed that genotypic CDTs include blaSME to promote the use of extended-spectrum cephalosporins when blaSME is detected.2 S. marcescens has been presumed to be at risk for the development of AmpC production, and thus cephalosporin resistance, although in vitro and clinical studies indicate clinically significant ampC expression in S. marcescens is unlikely.7 Therefore, while data are limited, it has been suggested extended-spectrum cephalosporins can be considered for treatment of SME-producing isolates if they test susceptible.2 Further, the clinical microbiology laboratory could append a comment that highlights the presence of a carbapenemase (SME)-producing isolate and counsels an infectious diseases consultation to determine the possibility of prescribing an extended-spectrum cephalosporin.
Clinical microbiology laboratories should have a high index of suspicion for SME-producing S. marcescens isolates when extendedspectrum cephalosporins test susceptible and carbapenems test resistant. For best practice, AST results should be confirmed and a test for enzymatic carbapenemase activity considered (currently, there are no US Food and Drug Administration-authorized diagnostic assays available that specifically identify SME). In our case, the laboratory’s algorithm is to reflex S. marcescens isolates that yield an AST profile suspicious for SME production immediately to a CDT. While awaiting confirmatory and supplemental test results, to avoid delays in reporting, clinical microbiology laboratories may want to report the isolate as a presumptive carbapenem-resistant Enterobacterales. For infection control and epidemiologic purposes, ruling out an OXA-48-like enzyme (which can generate a similar AST profile to SME producers) and other carbapenemases (IMP, KPC, NDM, and VIM) using a targeted genotypic or immunoassay could be performed.8,9 For SME-producing S. marcescens isolates, AST results should be reported as tested without modification of the interpretation of extended-spectrum cephalosporins.1 A comment recommending an infectious diseases consultation could be appended to the report. For those clinical microbiology laboratories that use cascade reporting protocols, unusual AST results, as observed in this case, must be confirmed and reported in full so providers do not assume that susceptibility to narrow-spectrum agents (eg, extended-spectrum cephalosporins) implies susceptibility to broader-spectrum agents (eg, carbapenems).1
In summary, SME-producing S. marcescens isolates can be presumed when an S. marcescens isolate tests susceptible to extendedspectrum cephalosporins and resistant to carbapenems, is negative for IMP, KPC, NDM, OXA-48-type, and VIM carbapenemases; and displays enzymatic carbapenemase activity. SME-producing isolates can be sent to a public health laboratory for further characterization, and many locales require that carbapenemase-producing Enterobacterales, including SME-producing S. marcescens, are submitted.
References
1. CLSI. Performance Standards for Antimicrobial Susceptibility Testing. 32nd ed. CLSI supplement M100. Malvern, PA: Clinical and Laboratory Standards Institute; 2022.
2. Bush K, Pannell M, Lock JL, et al. Detection systems for carbapenemase gene identification should include the SME serine carbapenemase. Int J Antimicrob Agents. 2013;41(1):1-4.
3. Bush K and Bradford PA. Epidemiology of β-lactamase-producing pathogens. Clin Microbiol Rev. 2020; 33(2):e00047-19.
4. Mataseje LF, Boyd DA, Delport J, et al. Serratia marcescens harboring SME-type class A carbapenemases in Canada and the presence of blaSME on a novel genomic island, SmarGI1-1. J Antimicrob Chemother. 2014;69(7):1825-1829.
5. Hopkins KL, Findlay J, Meunier D, et al. Serratia marcescens producing SME carbapenemases: an emerging resistance problem in the UK. J Antimicrob Chemother. 2017;72(5):1535-1537.
6. Pierce VM, Simner PJ, Lonsway DR, et al. Modified carbapenem inactivation method for phenotypic detection of carbapenemase production among Enterobacteriaceae. J Clin Microbiol. 2017;55(8):2321-2333.
7. Tamma PD, Aitken SL, Bonomo RA, et al. Infectious Diseases Society of America guidance on the treatment of AmpC β-lactamase-producing Enterobacterales, carbapenem-resistant Acinetobacter baumannii, and Stenotrophomonas maltophilia infections. https://www.idsociety.org/globalassets/idsa/practice-guidelines/amr-guidance/2.0/idsa-amr-guidance-v2.0.pdf. Accessed May 4, 2022.
8. Traczewski MM, Carretto E, Canton R, Moore NM, for the Carba-R Study Team. Multicenter evaluation of the Xpert Carba-R assay for the detection of carbapenemase genes in gram-negative isolates. J Clin Microbiol. 2018;56(8):e00272-18.
9. Jenkins S, Ledeboer NA, Westblade LF, et al. Evaluation of NG-Test Carba 5 for rapid phenotypic detection and differentiation of five common carbapenemase families: results of a multicenter clinical evaluation. J Clin Microbiol. 2020;58(7):e00344-20.
For more articles and issues of the AST News Update, click here.